5.2. Steps
You are welcome to use a dataset of your interest or alternatively download one from open TG-GATES, a large, publicly available, toxicological study from Japan: http://toxico.nibio.go.jp/open-tggates/search.html.
| i) | Select acetaminophen (or another chemical of interest) – search (use the English language version of the site, selector top right hand corner). |
| ii) |
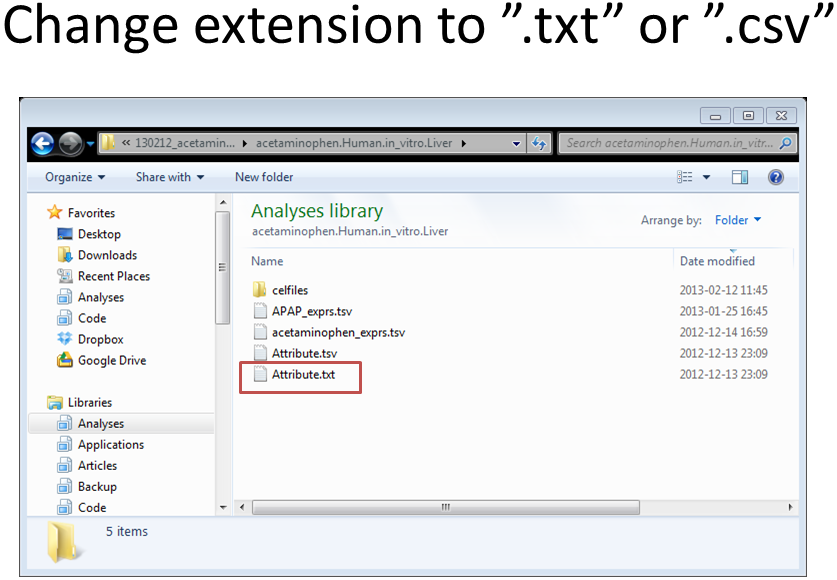
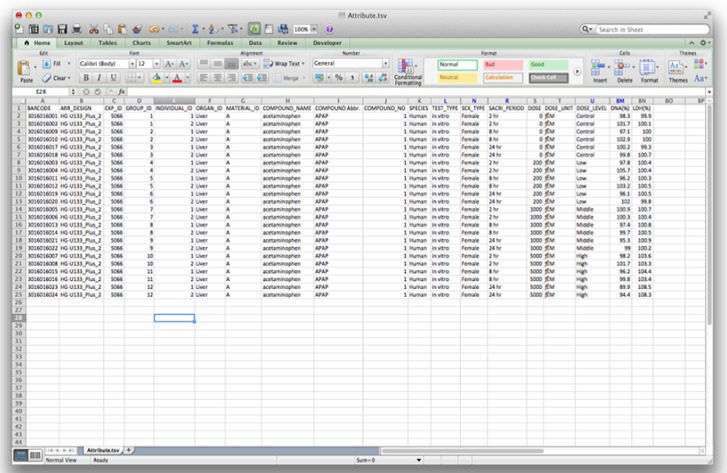
Select your organism (for example Human – in vitro – CEL files). Download all files: Extract the zip archive (for example acetaminophen.Human.in_vitro.Liver.zip). Rename “Attribute.tsv” to “Attribute.txt” and inspect in Excel. Make sure your file does not contain blank columns or special characters (such as μ). The labels in the “Attribute.txt” file are explained in the appendix (Chapter 6). |
| iii) | Data fields: |
| This is a dose-response study of acetaminophen. Fields are as follows: | |
| Barcode: Array Barcode Exp_ID: TGP internal experiment identifier |
| iv) | Start the SEURAT configured ISAcreator by double-clicking on the ISAcreator executable (ISAcreator.jar on PC, ISAcreator.app on the mac). |
Proceed to:
5.3. Step-by-step instructions for creating datasets (‘omics):
5.3. Mapping your annotation to ISA-tab, PART 1